The Husky Coronavirus Testing Program
Husky Coronavirus Testing has ended!
As of June 17th are no longer be handing out PCR swab kits or antigen tests. Antigen tests will still be available through UW. For more information, please visit the UW COVID-19 testing website
The Husky Coronavirus Testing program was a voluntary research study that ran from September 2020 to June 2023, providing COVID-19 testing for the university staff, faculty and students.
HCT initially grew out of a need for increased testing access at the beginning of the COVID-19 pandemic and has changed along the way, in partnership with the UW community. Over the last three years over 40,000 participants were enrolled, over 234,000 swabs were tested for SARS-CoV-2 and approximately 2,391,000 daily attestation surveys were completed by study participants.
Thus far, data from HCT has contributed to two published manuscripts and there is ongoing analysis of data from this study to inform scientific knowledge of rapid test accuracy, Long COVID in a university population, and vaccine effectiveness. HCT study research results are shown below and information about additional research manuscripts will be added as they are published and available to the public.
The university will continue to distribute COVID-19 antigen test kits free of charge. For more information, please visit the UW Covid-19 Testing website or UW EH&S Covid-19 Prevention and Response website.
Publications
Casto AM, Paredes MI, Bennett JC, Luiten KG, Han PD, Gamboa LS, McDermot E, Gottlieb GS, Acker Z, Lo NK, McDonald D, McCaffrey KM, Figgins MD, Lockwood CM, Shendure J, Uyeki TM, Starita LM, Bedford T, Chu HY, Weil AA. SARS-CoV-2 Diversity and Transmission on a University Campus across Two Academic Years during the Pandemic. Clin Chem. 2025 Jan 3;71(1):192-202. doi: 10.1093/clinchem/hvae194. PubMed PMID: 39749508.
Geng LN, Erlandson KM, Hornig M, Letts R, Selvaggi C, Ashktorab H, Atieh O, Bartram L, Brim H, Brosnahan SB, Brown J, Castro M, Charney A, Chen P, Deeks SG, Erdmann N, Flaherman VJ, Ghamloush MA, Goepfert P, Goldman JD, Han JE, Hess R, Hirshberg E, Hoover SE, Katz SD, Kelly JD, Klein JD, Krishnan JA, Lee-Iannotti J, Levitan EB, Marconi VC, Metz TD, Modes ME, Nikolich JŽ, Novak RM, Ofotokun I, Okumura MJ, Parthasarathy S, Patterson TF, Peluso MJ, Poppas A, Quintero Cardona O, Scott J, Shellito J, Sherif ZA, Singer NG, Taylor BS, Thaweethai T, Verduzco-Gutierrez M, Wisnivesky J, McComsey GA, Horwitz LI, Foulkes AS. 2024 Update of the RECOVER-Adult Long COVID Research Index. JAMA. 2024 Dec 18;. doi: 10.1001/jama.2024.24184. [Epub ahead of print] PubMed PMID: 39693079.
Kim AE, Bennett JC, Luiten K, O'Hanlon JA, Wolf CR, Magedson A, Han PD, Acker Z, Regelbrugge L, McCaffrey KM, Stone J, Reinhart D, Capodanno BJ, Morse SS, Bedford T, Englund JA, Boeckh M, Starita LM, Uyeki TM, Carone M, Weil A, Chu HY. Comparative Diagnostic Utility of SARS-CoV-2 Rapid Antigen and Molecular Testing in a Community Setting. J Infect Dis. 2024 Aug 16;230(2):363-373. doi: 10.1093/infdis/jiae150. PubMed PMID: 38531685.
Bennett JC, O'Hanlon J, Acker Z, Han PD, McDonald D, Wright T, Luiten KG, Regelbrugge L, McCaffrey KM, Pfau B, Wolf CR, Gottlieb GS, Hughes JP, Carone M, Starita LM, Chu HY, Weil AA. Evaluation of a novel university-based testing platform to increase access to SARS-CoV-2 testing during the COVID-19 pandemic in a cohort study. BMJ Open. 2024 Jun 4;14(6):e081837. doi: 10.1136/bmjopen-2023-081837. PubMed PMID: 38834321; PubMed Central PMCID: PMC11163660.
Kim AE, Bennett JC, Luiten K, O'Hanlon JA, Wolf CR, Magedson A, Han PD, Acker Z, Regelbrugge L, McCaffrey KM, Stone J, Reinhart D, Capodanno BJ, Morse SS, Bedford T, Englund JA, Boeckh M, Starita LM, Uyeki TM, Carone M, Weil A, Chu HY. Comparative diagnostic utility of SARS-CoV-2 rapid antigen and molecular testing in a community setting. J Infect Dis. 2024 Mar 26;. doi: 10.1093/infdis/jiae150. [Epub ahead of print] PubMed PMID: 38531685.
Bennett JC, Luiten KG, O'Hanlon J, Han PD, McDonald D, Wright T, Wolf CR, Lo NK, Acker Z, Regelbrugge L, McCaffrey KM, Pfau B, Stone J, Schwabe-Fry K, Lockwood CM, Guthrie BL, Gottlieb GS, Englund JA, Uyeki TM, Carone M, Starita LM, Weil AA, Chu HY. Utilizing a university testing program to estimate relative effectiveness of monovalent COVID-19 mRNA booster vaccine versus two-dose primary series against symptomatic SARS-CoV-2 infection. Vaccine. 2024 Feb 27;42(6):1332-1341. doi: 10.1016/j.vaccine.2024.01.080. Epub 2024 Feb 1. PubMed PMID: 38307746.
Pfau B, Opsahl J, Crew R, Best S, Han PD, Heidl S, McDermot E, Stone J, Schwabe-Fry K, MacMillan MP, O'Hanlon J, Sohlberg S, Acker Z, Ehmen B, Englund JA, Konnick EQ, Chu HY, Weil AA, Lockwood CM, Starita LM. Tiny swabs: nasal swabs integrated into tube caps facilitate large-scale self-collected SARS-CoV-2 testing. J Clin Microbiol. 2024 Feb 14;62(2):e0128523. doi: 10.1128/jcm.01285-23. Epub 2023 Dec 22. PubMed PMID: 38131692; PubMed Central PMCID: PMC10865831.
Weil AA, et al. Genomic surveillance of SARS-CoV-2 Omicron variants on a university campus. Nat Commun. 2022 Sep 6.
Weil AA, et al. SARS-CoV-2 Epidemiology on a Public University Campus in Washington State. Open Forum Infect Dis. 2021 Sep 17.
Media
Funding
Higher Education Emergency Relief Fund
Husky Coronavirus Testing Study Results Dashboard
As of May 2023
This section features a data dashboard summarizing results from Husky Coronavirus Testing research study and includes summaries and links to scientific publications utilizing HCT data. On behalf of the HCT study team, the BBI and the Chu lab, we would like to thank all of the individuals who have participated in the study for their time and contributions. This work would not be possible without you, and it is essential to understanding and combating COVID-19 and keeping the UW community safe and informed.
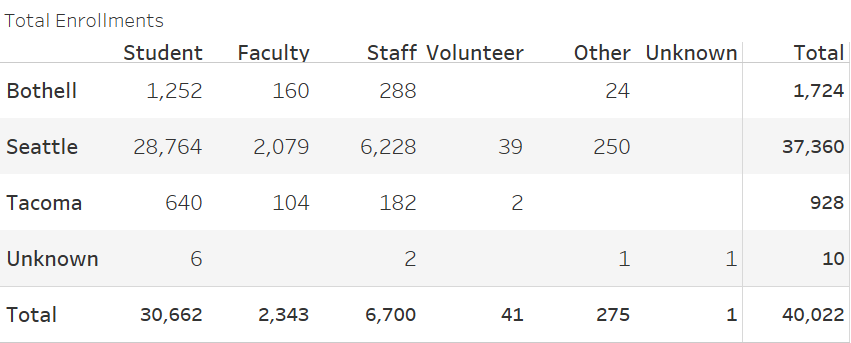
This figure shows the total enrollments in HCT by campus and participant type.
This figure shows a breakdown of the age of all HCT participants.
This figure shows a breakdown of the race of all HCT participants.
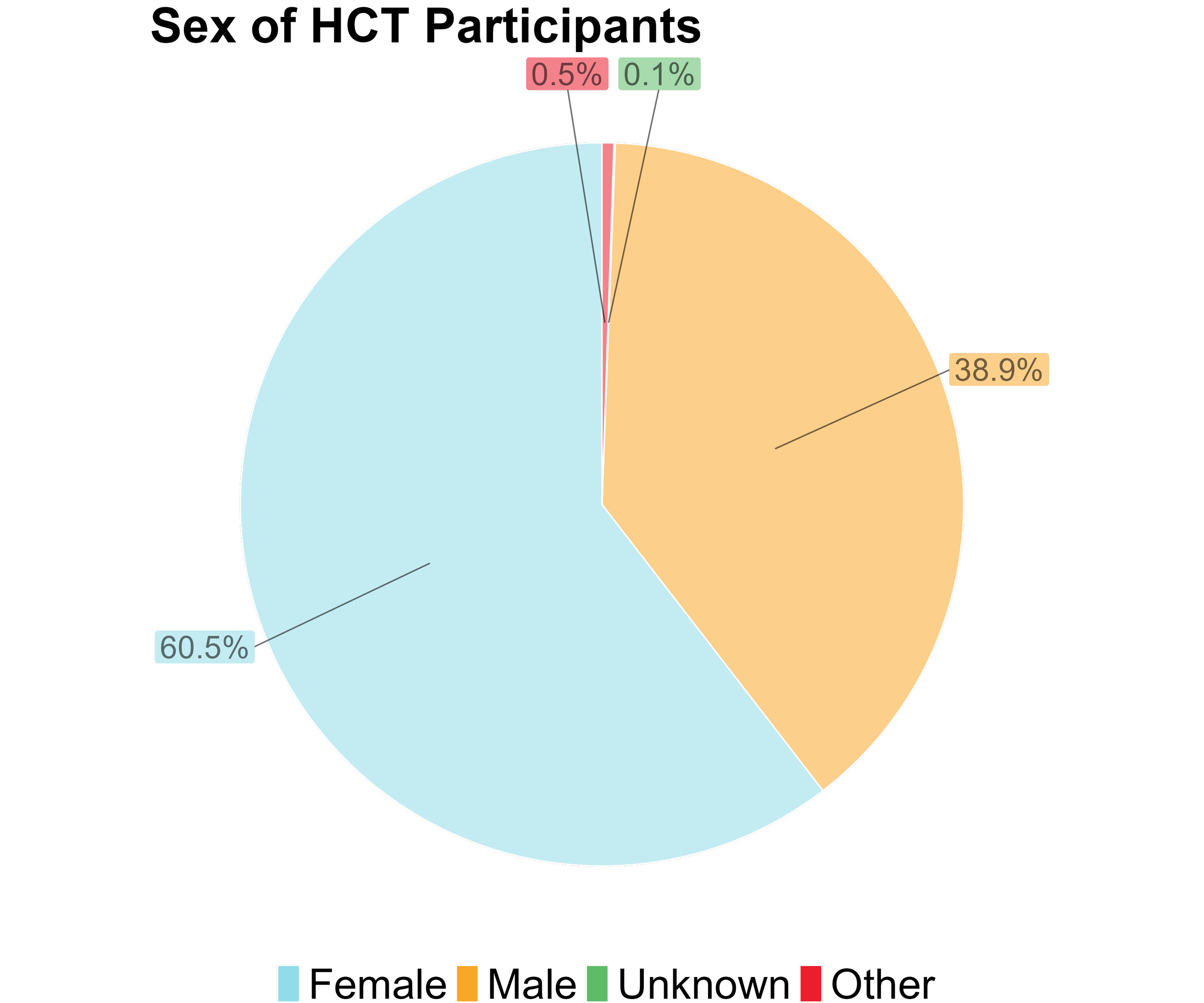
This figure shows a breakdown of the sex of all HCT participants.
This image displays the daily PCR tests, positive PCR tests and percent positivity of COVID-19 from September 2020 to the present. The pink bars represent total daily tests, which peaked at about 1500 in a single day during the omicron surge in January of 2022. The regular decreases down to approx 100 tests per day represent weekends, where testing interest decreases significantly. The green bars represent individual positive and inconclusive cases each day and the blue line shows this as a percent of the total tests. The dramatic increase in positivity as of summer 2022 may be due to less participants utilizing PCR tests in general with the widespread availability of rapid testing or participants utilizing PCR tests specifically to confirm their positive rapid tests.
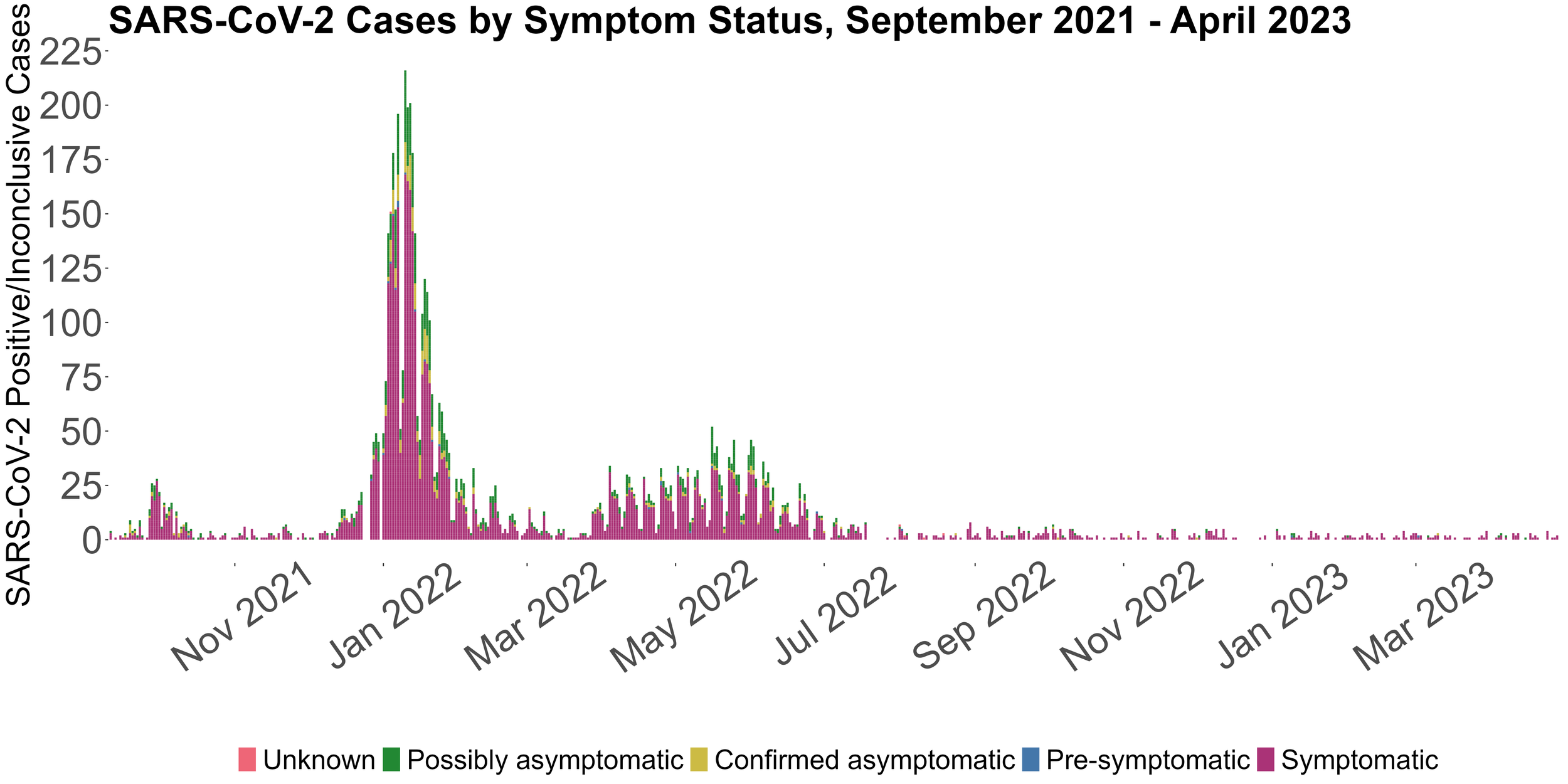
This image displays the daily positive COVID-19 PCR tests from Fall 2021 to the present labeled by symptom status.
This image displays the daily positive flu PCR tests from January 2023 to the end of April. Due to technical issues and low levels of cases circulating in the community, HCT has paused testing samples for influenza.
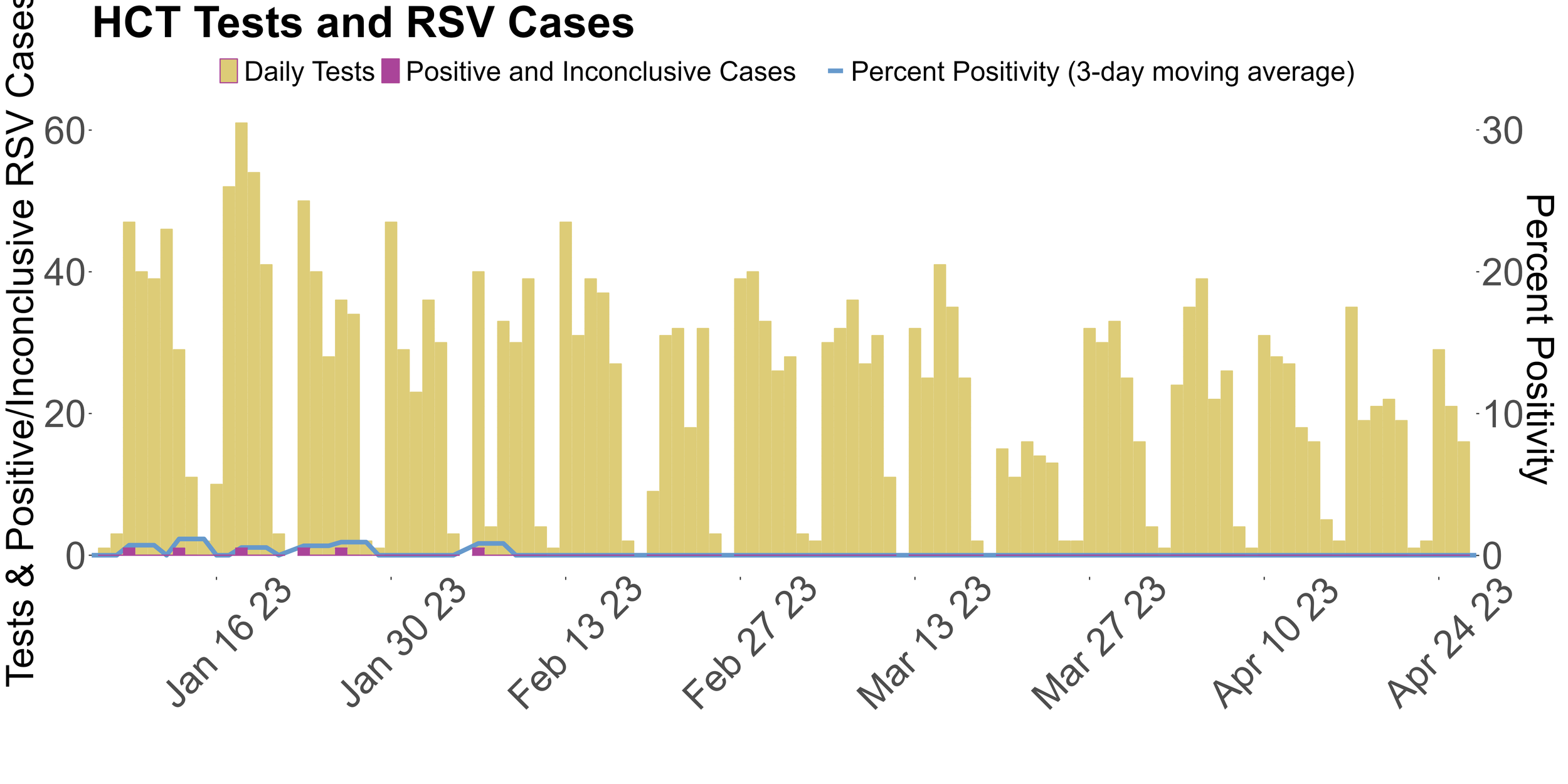
This image displays the daily positive RSV PCR tests from January 2023 to the end of April. Due to technical issues and low levels of cases circulating in the community, HCT has paused testing samples for RSV.
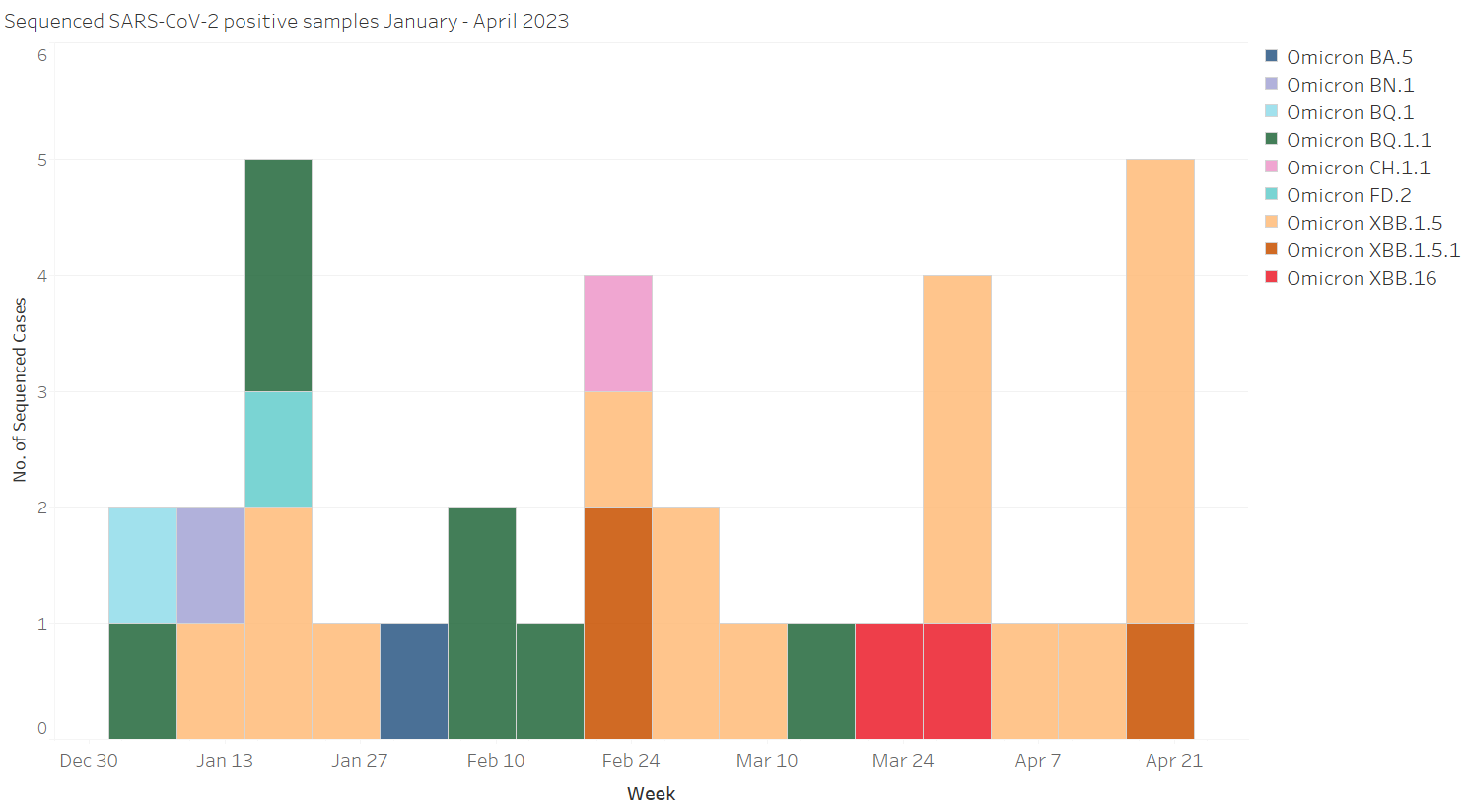
This image shows the prevalence of SARS-CoV-2 variants from September of 2020 to the present. Each color represents a different variant and is measured by individual sequenced cases over time.
This figure shows the current average prevalence of SARS-CoV-2 variants, with each color representing the percentage of the sequenced positive HCT swabs identified as each variant. Sequencing takes approx 2-4 weeks to complete and report after a swab is tested.
This figure shows the total number of rapid test results reported daily (orange), the number of positive rapid test results reported daily (blue) and the percent positivity of the reported rapid tests (green).
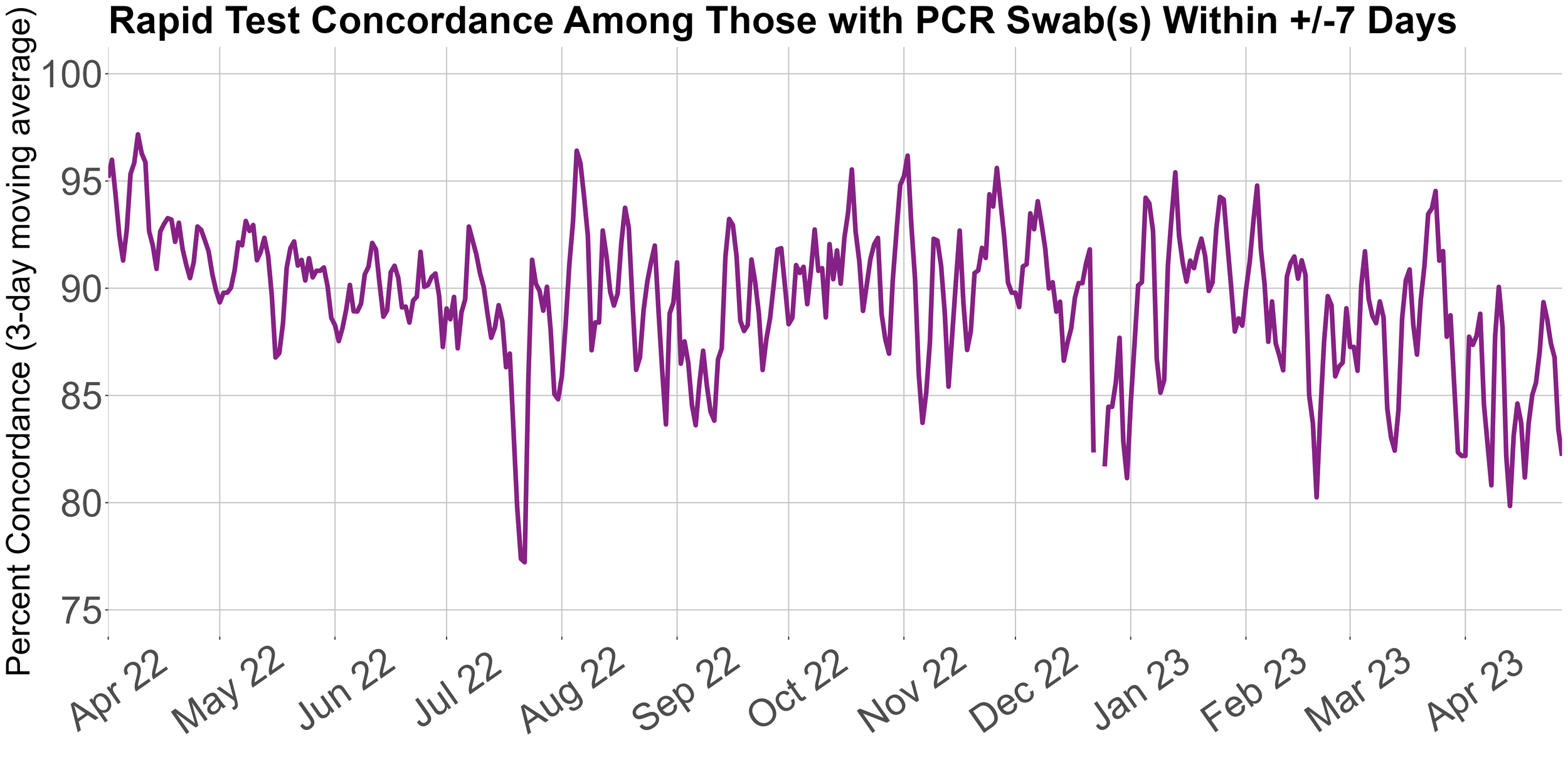
This figure displays the concordance over time between positive rapid test results reported to HCT and matching PCR test results within one week of the original positive rapid test.
Current Husky Coronavirus Testing Study Publications
As of May 2023
Genomic surveillance of SARS-CoV-2 Omicron variants on a university campus
Nature Communications, 09/06/2022
This paper presents the results of genomic surveillance of SARS-CoV-2 Omicron variants on the University of Washington campus. The authors aimed to understand the spread and prevalence of Omicron variants in a localized setting and its impact on the community. The study was conducted over the course of two years at the University of Washington, where weekly surveillance testing was conducted for students and staff. Samples that tested positive for SARS-CoV-2 were sequenced to determine the variant.
The study found that total of 3,048 of 24,393 individuals tested positive for SARS-CoV-2 by RT-PCR; whole genome sequencing identified 209 Delta and 1,730 Omicron genomes of the 1,939 total sequenced. Compared to Delta, Omicron had a shorter median serial interval between genetically identical, symptomatic infections within households (2 versus 6 days, P=0.021). Omicron also demonstrated a greater peak reproductive number (2.4 versus 1.8) and a 1.07 (95% confidence interval: 0.58, 1.57; P<0.0001) higher mean cycle threshold value.
Despite near universal vaccination and stringent mitigation measures, Omicron rapidly displaced the Delta variant to become the predominant viral strain and led to a surge in cases in a university population in January 2022. The authors also found evidence of intra-host diversity within Omicron variants, suggesting ongoing evolution of the virus within individuals. The study highlights the importance of genomic surveillance to track the emergence and spread of variants, and to inform public health measures.
The authors conclude that their findings provide important insights into the epidemiology and dynamics of Omicron variants in a localized setting. The study emphasizes the need for continued genomic surveillance to monitor the evolution and spread of SARS-CoV-2 variants and to inform public health interventions. The study also highlights the importance of implementing measures to prevent transmission, such as vaccination, masking, and social distancing, to limit the impact of the pandemic on communities.
SARS-CoV-2 Epidemiology on a Public University Campus in Washington State
Open Forum Infectious Diseases, 09/17/2021
This publication aimed to evaluate the Husky Coronavirus Testing program’s impact of SARS-CoV-2 transmission on the University of Washington campus and measure spread in the university community using viral genome sequencing.
HCT uses remote contactless enrollment, daily mobile symptom and exposure tracking, and self-swab sample collection. Individuals were tested if the participant was exposed to a known SARS-CoV-2-infected person, developed new symptoms, or reported high-risk behavior (such as attending an indoor gathering without masking or social distancing), if a member of a group experiencing an outbreak, or at enrollment. Study participants included students, staff, and faculty at UW during the Autumn quarter of 2020.
In the fall of 2020 HCT enrolled 16,476 individuals, performed 29,783 SARS-CoV-2 tests, and detected 236 infections. Seventy-five percent of positive cases reported at least 1 of the following: symptoms (60.8%), exposure (34.7%), or high-risk behaviors (21.5%). Greek community affiliation was the strongest risk factor for testing positive, and molecular epidemiology results suggest that specific large gatherings were responsible for several outbreaks.
The paper concludes a testing program focused on individuals with symptoms and unvaccinated persons who participate in large campus gatherings may be effective as part of a comprehensive university-wide mitigation strategy to control the spread of SARS-CoV-2.
This figure shows a phylogenetic tree of SARS-CoV-2 samples from Washington, including 88 samples from this study. Included here are all SARS-CoV-2 genomes from Washington collected on or after September 25, 2020, a random subsample of 1000 Washington samples collected before September 25, and the Wuhan/Hu-1 reference genome. Samples are positioned on the x-axis by date of collection. The indicated groupings show a large outbreak within the Greek community distinct from the outbreaks in the local population at large.