Hospitalized or Ambulatory Adults with Respiratory Viral Infections
Our group has a long-standing interest in respiratory syncytial virus immune profiling studies and development of RSV vaccines and has a cohort study of adults with respiratory viral infections, entitled HAARVI. In the last four years, we utilized this infrastructure to enroll individuals recovered from SARS-CoV-2 infection into a prospective cohort study, including the first US patient, now followed out to three years, and have utilized this cohort to collect the clinical samples for many basic science investigators in human immunology and virology. This cohort has been foundational for much of the understanding around durability of immunity, and ability to evade novel variants. We provided samples from the first US patients with SARS-CoV-2 infection to multiple groups early in the pandemic, including the NIH and DARPA, and these samples permitted identification of several potent monoclonal antibodies, including the Eli Lilly monoclonal antibody in use, and also provided a correlate of protection for the early Moderna mRNA 1273 vaccine trials.
We have multiple ongoing collaborators at Vanderbilt, Ragon Institute of Harvard-MIT, Emory University, Institute for Systems Biology, Fred Hutchinson, Institute for Systems Biology, and the University of Washington. These studies have been important in answering the virologic and immunologic questions around hybrid immunity, vaccine breakthrough, variant response, and predictors of long COVID.
Media
https://www.nytimes.com/2022/01/25/health/long-covid-risk-factors.html
https://newsroom.uw.edu/news/breakthrough-infections-spur-strong-antibody-responses%C2%A0
https://www.cnn.com/2021/02/19/health/post-covid-syndrome-long-haulers-gupta-wellness/index.html
https://newsroom.uw.edu/news/%E2%80%8Beven-mild-cases-covid-19-impact-can-persist
Funding
Gates Foundation, DARPA, NIAID
Current HAARVI publications
NCBI HAARVI Publication List
This section features summaries and links to scientific publications the HAARVI research study samples and data have contributed to. On behalf of the HAARVI study team and the Chu lab, we would like to thank all of the individuals who have participated in the study for their time and contributions. This work would not be possible without you, and it is essential to understanding and combating COVID-19 and the global pandemic.
If you have recently tested positive for Flu or RSV in the Seattle area and would like to enroll in our study, please fill out our interest form here: http://tinyurl.com/UWCOVID
Keywords:
Antibody/Antibodies: Small proteins that are produced by the immune system in response to specific pathogens such as viruses and harmful bacteria. These proteins are the body’s natural defense system against infection and can protect against future illness from the same pathogen.
Antigen (Ag): A molecule or molecular structure on a pathogen that can be bound by an antibody and induce an immune response in the body. Antibodies can be produced to match and bind to specific antigens and allow the body to “remember” that virus or bacteria in the future. Vaccines often use antigens to trigger the production of antibodies against a specific pathogen without causing an active infection.
B cells: A type of white blood cell that produces antibodies and aids in the detection and identification of pathogens in the body.
Epitope: The specific site or part of an antigen where an antibody binds to. On larger proteins and molecules, antibodies are only big enough to interact with the epitope structure due to its small size.
Monoclonal antibodies (mAbs): A type of antibody produced as clones of a single parent antibody that targets and binds to a specific protein or antigen in the body, such as cancer cells or pathogens, for a variety of effects.
Monoclonal Antibody Therapy: A form of immunotherapy where patients are administered synthetic monoclonal antibodies that trigger an immune response to a specific pathogen or cell. This prompts the immune system to begin creating antibodies that fight the infection. The advantages of monoclonal antibody therapeutics are the antibodies can easily be manufactured in large quantities, side effects are generally less severe than treatments such as radiation and the timelines for the development, testing, and approval are generally shorter than other treatments such as vaccines, antivirals or steroids. Many current research studies are focused on discovering and reproducing monoclonal antibodies that specifically target the SARS-CoV-2 virus.
Neutralizing antibodies: A type of antibody that defends a healthy cell by rendering the virus non-infectious through various mechanisms.
Nucleoprotein: A type of protein structurally associated with DNA or RNA.
Pathogen: A broad term for any microorganism that can cause diseases, including viruses and bacteria.
SARS-CoV-2: The name of the specific coronavirus that led to the global pandemic starting in March of 2020. This virus causes the disease COVID-19, although the two terms are often used interchangeably by the media.
Spike protein: A type of protein on the surface of the SARS-CoV-2 virus that facilitates the entry of the virus into host cells (such as lung cells) by binding to a receptor on the host cell surface. There is evidence some antibodies may detect or neutralize the virus through this protein. These proteins give the virus its distinct “crown” that it’s named for.
T Cell: A class of white blood cells imperative to the functioning of the immune system. There are multiple types, the most commonly discussed being “helper” T cells (CD4+) and “killer” or cytotoxic T cells (CD8+). They are responsible for activating the humoral immune response (production of antibodies) and killing cells that have been infected with a virus.
Viral load: A numerical expression of the quantity of virus in a given volume of fluid, such as sputum or blood.
Published in Journal of Immunology on 5/1/2023
This study was conducted in coordination with the Seshadri Lab to better understand why people who had COVID-19 before receiving an mRNA vaccine were better protected from getting the virus again, compared to those who were only vaccinated - a phenomenon known as hybrid immunity. Subjects for this investigation were members enrolled in the HAARVI study.
CD4 T cells are a type of white blood cell responsible for signaling to other immune cells to fight a virus. These cells include B-lymphocytes (cells that create antibodies), and cytotoxic T cells, such as CD8 T Lymphocytes, which are crucial in killing virus infected cells. Researchers found similar levels of overall immune response after vaccination in both groups, but those who previously had COVID-19 did have a stronger response from CD4 T cells. They also found that CD8 T cells were more effective in targeting the virus in those who had COVID-19 before.
These results suggest that having COVID-19 before getting vaccinated may have improved the immune system’s ability to fight the virus when it was encountered again. Future areas of research may investigate other types of acquired hybrid immunity, for instance the immune response of vaccinated individuals who are then infected with SARS-CoV-2.
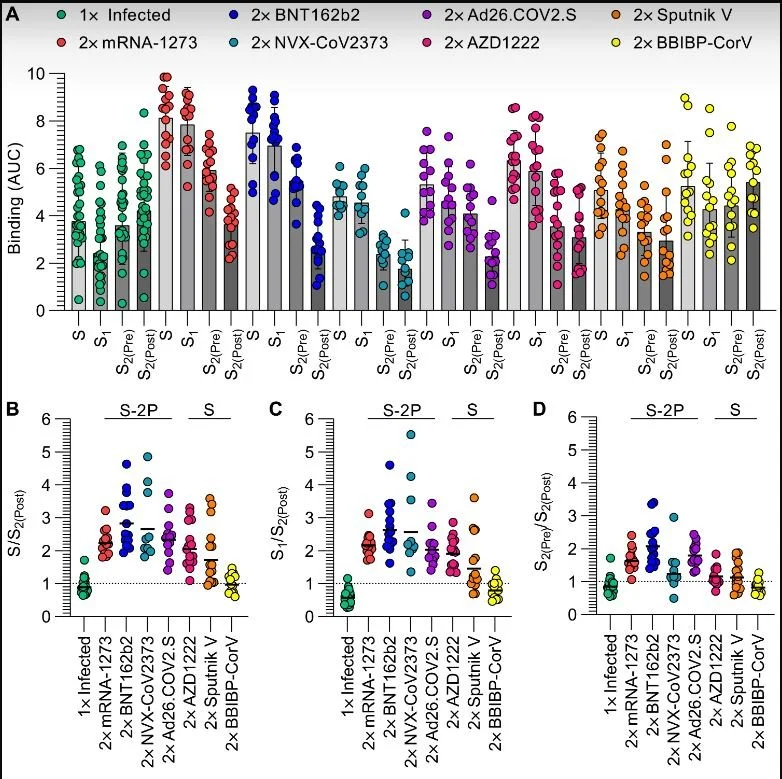
SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines
Published in Science Immunology on 11/10/2022
The Veelser Lab at the University of Washington studies the structural biology of SARS-CoV-2 and how the immune system fights the virus. This study used a variety of methods to look at where the antibodies made by our immune system target the spike protein of SARS-CoV-2. Antibodies from individuals immunized with one of seven different types of SARS-CoV-2 vaccines were compared, and it was confirmed that not stabilizing a specific region of spike called the S2 domain when designing vaccines led to worse viral neutralization. This study confirms the importance of considering the structural biology of both vaccines and the immune response they elicit when designing vaccines. c
Estimated Global Proportions of Individuals With Persistent Fatigue, Cognitive, and Respiratory Symptom Clusters Following Symptomatic COVID-19 in 2020 and 2021
Published in JAMA on 10/10/2022
There are many challenges in accurately estimating the number of people who experience “long-COVID”, now termed post-COVID conditions (PCC). Studies use different surveys, methods, symptom lists, metrics and populations to ask about PCC, which has produced widely varying estimates. This study performed by the Institute for Health Metrics and Evaluation (IHME) at UW looks at data from multiple cohorts, studies, and health systems to estimate an overall prevalence of PCC. They grouped conditions experienced after COVID-19 into three categories: persistent fatigue with bodily pain or mood swings, cognitive problems, and ongoing respiratory problems. Their modeling estimates that 6.2% of individuals experience conditions in at least one of the three symptom categories at 3 months after symptomatic SARS-Cov-2 infection. They also found that women and those who had severe illness requiring hospitalization were more likely to have PCC.
Receptor-Binding Domain (RBD) Antibodies Contribute More to SARS-CoV-2 Neutralization When Target Cells Express High Levels of ACE2
Published in Viruses on 9/16/2022
Neutralization assays are a key tool in understanding how our immune systems kill SARS-CoV-2. This paper systematically compares and describes the differences in cell types commonly used for these assays. Serum from HAARVI participants was used as a standard in these assays.
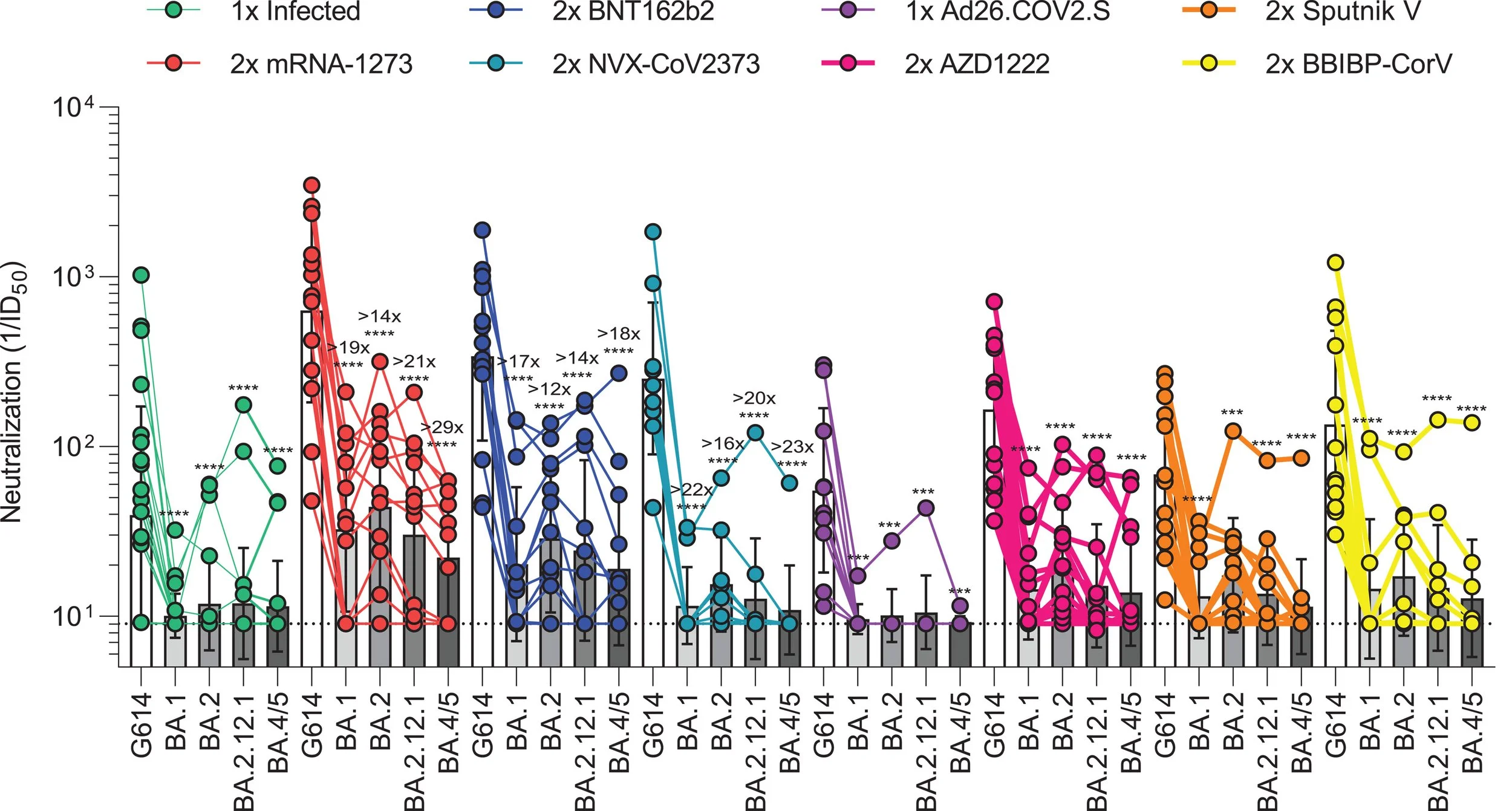
Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines
Published in Science on 07/19/2022
This study tests the immunity elicited by a number of COVID-19 vaccines and how well they neutralize or kill the different omicron variants. Overall, the omicron variants were able to escape pre-existing immunity, however, receiving booster doses provides better protection than the primary series alone.
Neutralization of individuals who received different vaccine types. A higher the neutralization number means better killing or the virus.
Challenges and lessons in establishing human immune profiling cohort studies for pandemic response
Published in Immunological Reviews on 6/30/2022
This review written by HAARVI project manager, Jenni Logue, and principal investigator, Dr. Helen Chu, details the challenges and importance of launching the HAARVI study at the beginning of the COVID-19 pandemic. It discusses regulatory barriers faced at the start of the pandemic, lessons learned for future pandemics, and the importance of cohort studies for pandemic preparedness.
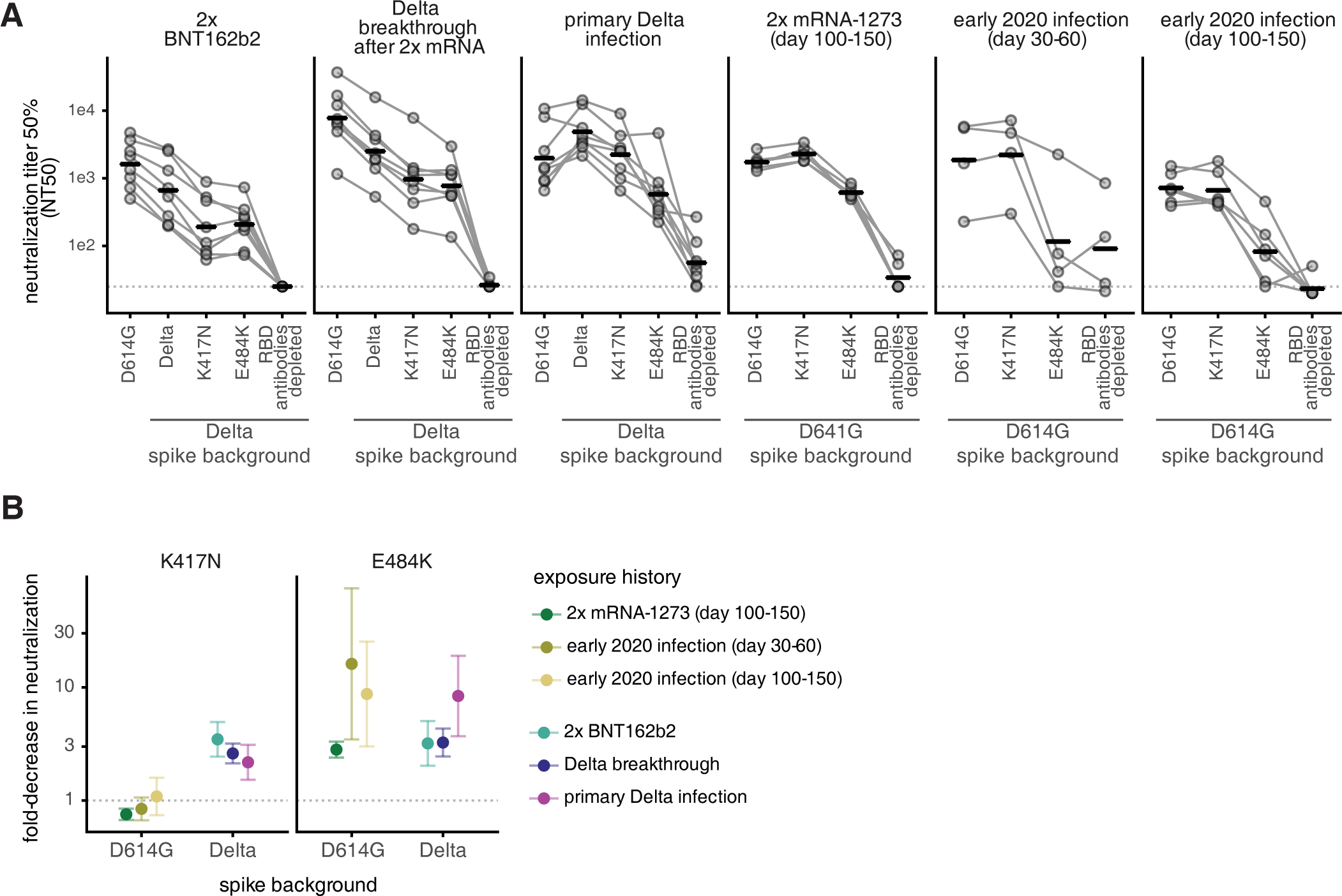
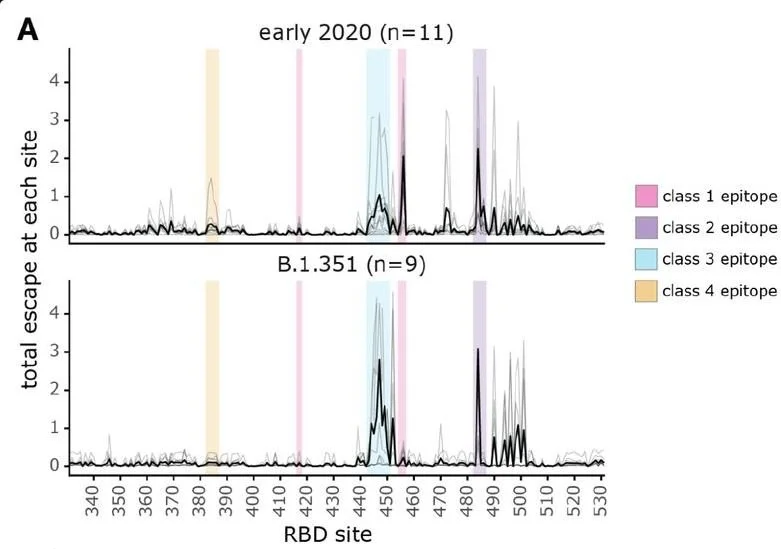
The SARS-CoV-2 Delta variant induces an antibody response largely focused on class 1 and 2 antibody epitopes
Published in PLOS Pathogens on 06/29/2022
In this study, conducted by the Bloom Lab at the Fred Hutchinson Cancer Center, HAARVI samples were used to characterize where the antibodies made by our immune systems bind to the SARS-CoV-2 viral particle. As the SARS-CoV-2 virus has continued to quickly mutate into new variants, the concept of antibody escape has become increasingly important. Mutations in the virus allow new variants to escape our existing immunity that comes from vaccination, previous infection, or a combination of both called “hybrid” immunity. Using a system called deep mutational scanning, the Bloom lab can create yeast cells that express versions of the spike protein with mutations in single amino acids. This is done for every possible amino acid position on the spike protein. They then test to see which mutations are able to escape previous immunity. Interestingly, one mutation that is found in many of the omicron variants was found to be particularly prone to immune escape.
Understanding how viruses may evolve in the future to escape our immune systems has important implications for vaccine and drug development. In the future, we may be able to use this information to protect against future viral variants more effectively.
Figure A shows how well antibodies from each of the groups listed on the top row can neutralize or kill the virus. The Y axis shows neutralization, a higher number indicates the antibodies are better at killing COVID-19. The X axis shows which variant of the virus was used in the assay. For example, in the first group that received 2 doses of the Pfizer vaccine (BNT162b2), their antibodies neutralize the early Wuhan strain well, but were worse at neutralizing the Delta strain the virus with the K417N or E484K mutations.
Multi-site observational maternal and infant COVID-19 vaccine study (MOMI-vax): a study protocol
Published in BMC Pregnancy and Childbirth on 5/12/2022
This paper details the protocol for a national, multi-site study that assesses the safety and effectiveness of COVID-19 vaccines in pregnant and lactating women. This is an ongoing study that is expected to help inform personal decision-making and policy recommendations regarding the use of approved COVID-19 vaccines in pregnant and postpartum individuals. HAARVI sera and breastmilk were used to validate the assays that will be used to measure neutralizing antibodies in study participants.
Imprinted SAS-CoV-2-specific memory lymphocytes define hybrid immunity
Published in Cell on 04/28/2022
Understanding the differences between COVID-19 infection, vaccination, and a combination of the two, are of the utmost relevance as a majority of the US has now been vaccinated. This study, led by the lab of Marion Pepper at the University of Washington, examines the difference in immune response between those who have been vaccinated only, and those who have been infected then vaccinated which is also called “hybrid” immunity. Using samples from the HAARVI study, they showed that vaccination creates a robust immune memory in both the vaccinated and hybrid cohort. However, there were certain characteristics of the hybrid group that were not seen in the vaccine only group, like the production certain signaling molecules called cytokines that help our body clear infection. Further research will be needed to understand the long-term impact of multiple vaccine doses and hybrid immunity.
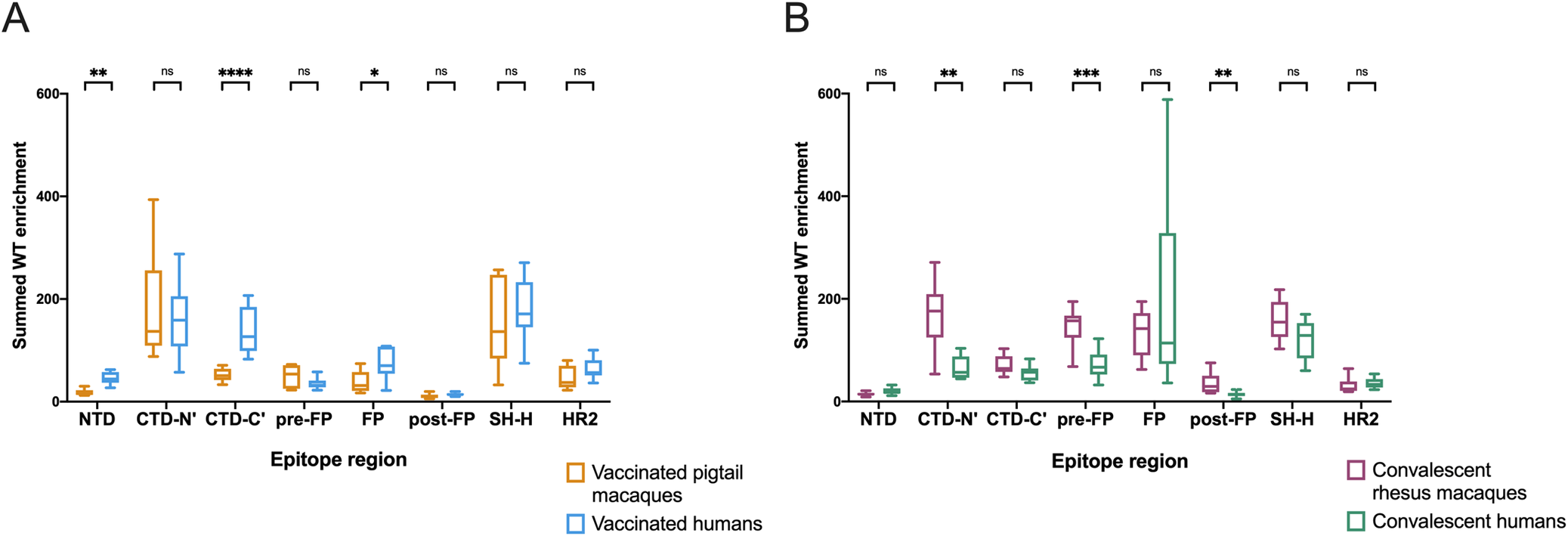
Detailed analysis of antibody responses to SARS-CoV-2 vaccination and infection in macaques
Published in PLOS Pathogens on 4/11/22
Macaque monkeys are commonly used in biomedical research, especially those focused on immunology and infectious diseases, as a proxy for human responses. However, small differences exist between the human and macaque immune response; therefore, it is important to quantify how closely these monkeys model human infection and vaccination responses. This study focused on comparing immune responses in mRNA vaccinated and convalescent (post-infection) humans against macaques challenged with the SARS-CoV-2 virus. The receptor-binding domain (RBD) was not detailed since it has been extensively researched and the screening methods of this study did not allow for detection of conformational epitopes that occur when the protein is properly folded; instead, the researchers focused on linear epitopes depicted in the amino acid sequences themselves. The researchers found that while macaque models produced similar immunity as human infections, there were distinctions between the two species in both vaccinated and convalescent groups regarding which epitopes were preferentially bound as well as individual variation in the same realm. The researchers also quantified differences in escape pathways, mutations within the virus that allow it to avoid binding with host antibodies, between vaccinated and convalescent humans and macaques. Overall, macaques model human SARS-CoV-2 infections in the recognition of many major epitopes but antibody responses differ to a degree that may warrant further investigation into the full spectrum of variations between the two groups.
This image depicts the differences between groups in wildtype peptides within each epitope region.
SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses
Published in Cell on 3/3/2022
The delta variant of SARS-CoV-2 was dominant in the US from late June of 2021 until the emergence of the Omicron variant in December 2021. This study, led by the Veesler lab at the University of Washington, looked at the antibodies produced by individuals with breakthrough infections (BTI) and compared them to those who had received mRNA COVID-19 vaccines only. They found that those with BTI had a stronger and more durable immune response than those who only received the 2-dose primary series of mRNA vaccines. However, a third dose of mRNA vaccine led to a comparable antibody response in the vaccine only group to those with BTI and improved the immune response to the Omicron variant.
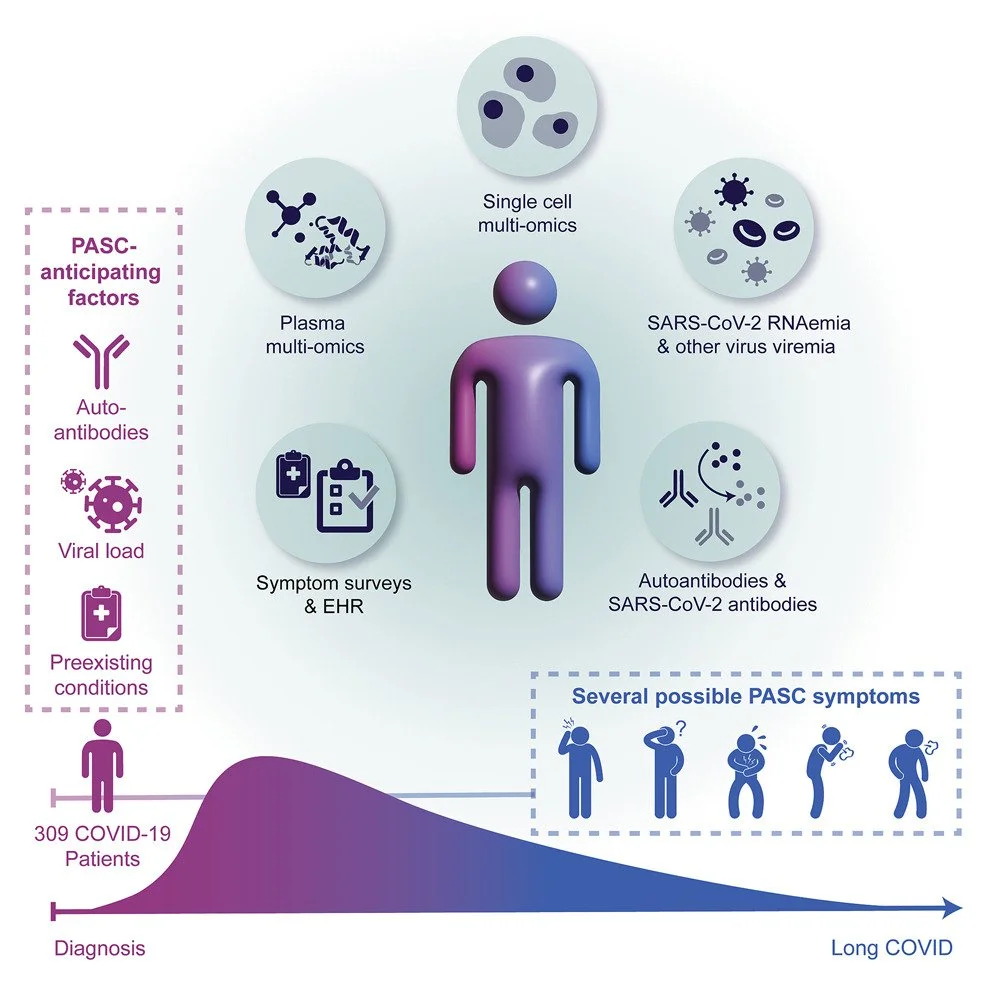
Multiple early factors anticipate post-acute COVID-19 sequelae
Published in Cell on 01/24/2022
This extremely comprehensive study examines multiple factors to predict post-COVID-19 conditions (PCC) or “long-covid”. The study was led by Dr. James Heath at the Institute for Systems Biology (ISB) and was made in collaboration with Dr. Jason Goldman at Swedish Medical Center (SMC). Samples provided from the HAARVI study and the INCOV cohort study at SMC were tested to examine immune cells, antibodies, viral load, multi-omics and health data. These elements were brought together in a large-scale analysis to determine factors that increased the risk of developing PCC. The factors identified were the presence of autoantibodies (antibodies that mistakenly target proteins or molecules that are part of our bodies), high SARS-CoV-2 viral load during acute illness, pre-existing type 2 diabetes, and Epstein-Barr Virus viremia (the presence of virus in the blood).
The Chu Lab has continued collaboration with the ISB and SMC as part of the NIH reCOVer study on PCC and additional projects. More information on the recover study can be found at https://isbscience.org/pnwrecover/
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection
Published in eLife on 1/24/2022
The spike protein of SARS-CoV-2 is an important target for our immune system to effectively inactivate and neutralize the virus. If the spike protein is blocked by an antibody, the virus cannot gain entry or interact with human cells. This paper explores where antibodies bind the spike protein in individuals who were vaccinated compared to those who were infected and unvaccinated at the time of their illness. They found that SARS-CoV-2 mRNA vaccination resulted in antibodies binding to more locations on the spike protein than infection only. This suggests that protection could vary depending on the route of exposure to spike antigen. However, future variants of SARS-CoV-2 may be able to easily escape vaccine induced immunity and considerations need to be taken when designing future vaccines.
Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift
Published in Nature on 12/23/21
The omicron variant (B.1.1.529) of SARS-CoV-2 has rapidly spread to become the dominant strain in many countries around the world. A unique combination of mutations have allowed it to more effectively infect people, even those who already have antibodies against SARS-CoV-2 either from vaccination or previous infection. This thorough analysis characterizes these mutations and tested how well antibodies are able to neutralize or stop the omicron variant. As the figure below indicates, there is a large decline in effectiveness against omicron in those who have been previously vaccinated and/or infected. The largest drop was in individuals who received adenovirus vaccines, such as the Johnson & Johnson and Astrazeneca, while the best antibody response was found in those with both a previous infection and two doses of an mRNA vaccine. This is an important demonstration of vaccine efficacy against the omicron variant, showing that being vaccinated with an mRNA vaccine provided better protection than previous infection alone.
The Receptor Binding Domain (RBD) on the spike protein, is a target of many monoclonal antibody (mAb) drugs. If the RBD site is blocked, the spike protein on the virus cannot connect to human cells and infect them. Mutations in the spike protein of the omicron variant have prevented many mAbs from binding to it, rendering them effectively useless. This study tested many mAb drugs and combinations of these drugs to see which still remain effective against omicron. This provides strong potential clinical guidance on which drug(s) to use as the omicron variant continues to spread.
Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants
Published in Science on 11/9/2021
Widespread transmission of SARS-CoV-2 has led to the emergence of new viral strains due to mutations in the spike protein of the virus. While several different variants have arisen, the delta variant of SARS-CoV-2 has quickly spread around the world due to increased transmissibility. This study, conducted by the Veesler Lab at the University of Washington, looked at how antibodies produced by individuals who received different SARS-CoV-2 vaccines neutralized the delta, delta+ and kappa variants of SARS-CoV-2. When compared to the original strain of the virus, they found that the variants were able to more effectively evade neutralization by vaccine-elicited antibodies in people immunized with the Moderna, Pfizer and J&J vaccines. The research also identified a specific mutation in the kappa and delta variants that decreased neutralization. This knowledge may aid in the development of vaccines that will be more effective against emerging strains of SARS-CoV-2.
DISCLAIMER: studies posted to medRxiv or biorxiv (med archive and bio archive) are pre-prints and have not been peer reviewed or published.
The figure on the left shows the neutralization levels of the viruses (A-C) and incidence of the variants (D-F). Using a technique called CryoEM, the team is able to look at detailed structural changes to the spike proteins in these variants (shown on the right).
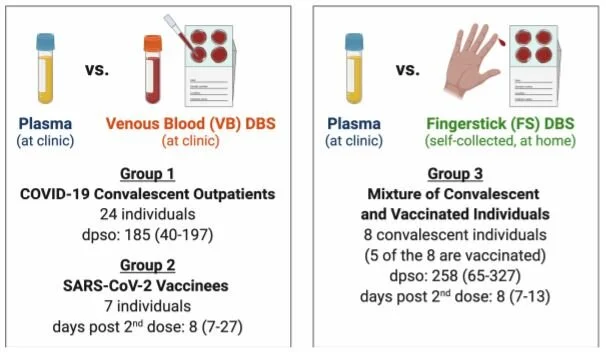
SARS-CoV-2 antibody binding and neutralization in dried blood spot eluates and paired plasma
Published in Microbiology Spectrum 10/20/2021
HAARVI participants self-collected dried blood spots at home via fingerstick in this pilot study to assess the utility and feasibility of using dried blood spots (DBS) for detection and characterization of SARS-CoV-2 antibodies. These self-collected samples were compared to plasma samples collected from the same individuals via traditional blood draw (venipuncture by a trained phlebotomist). Multiple analyses were carried out to compare antibody levels and responses in the two sample types, and ultimately showed a strong agreement between the paired DBS and plasma samples. This study has shown that DBS cards can be an effective and accurate way to collect and study serological samples. DBS cards have numerous strengths including their low cost and stability and may be a simple, cost-effective sample collection method for future SARS-CoV-2 studies.
Thank you to the HAARVI participants who participated in this pilot study!
DISCLAIMER: studies posted to medRxiv or biorxiv (med archive and bio archive) are pre-prints and have not been peer reviewed or published.
Characteristics of the paired sample groups
A SARS-CoV-2 variant elicits an antibody response with a shifted immunodominance hierarchy
In preprint on biorxiv on 10/13/2021
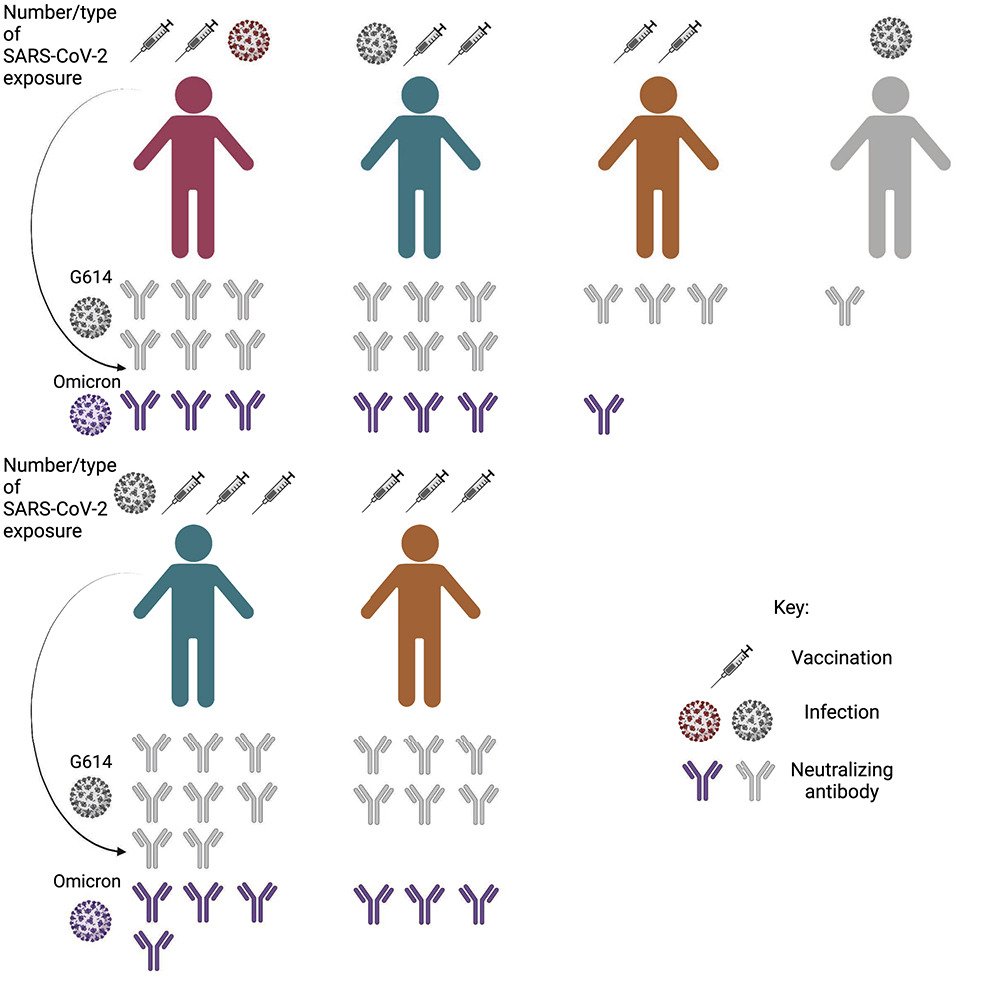
SARS-CoV-2 variants have mutations at key sites (epitopes on the spike protein/receptor binding domain) targeted by antibodies. These key sites have immunodominance hierarchies, differences in epitopes targeted by antibodies. While SARS-CoV-2 isolates from early 2020 are neutralized via their class 1 and 2 epitopes, antibodies neutralize B.1.351 (beta) variants mainly by class 3 epitopes.
Overtime, the immunodominance hierarchies shifted. Individuals will accumulate increasingly different exposure histories through infection and vaccination. This causes different antibody neutralization range and specificity. In the figure, the total escape rate is the rate at which mutations in these sites evade the immune antibody response. Class 1 and 2 epitopes in the early 2020 isolates have the greatest escape rate while class 2 and 3 have the greatest escape rate in beta variants. This means class 1 & 2 epitopes are major targets of the antibody response against early 2020 isolates while class 3 epitope-targeted antibodies are more significant in neutralization of beta variants than early 2020 isolates. These results indicate immunodominance hierarchies shifting from the immunodominant class 1 and 2 epitopes of SARS-CoV-2 to the class 3 epitope.
Such shifts can have important implications and tell us that individuals who have been exposed to different strains of the virus may have different susceptibilities to other SARS-CoV-2 variants. Understanding the immunity developed after exposure to different SARS-CoV-2 variants is important as the virus continues to evolve.
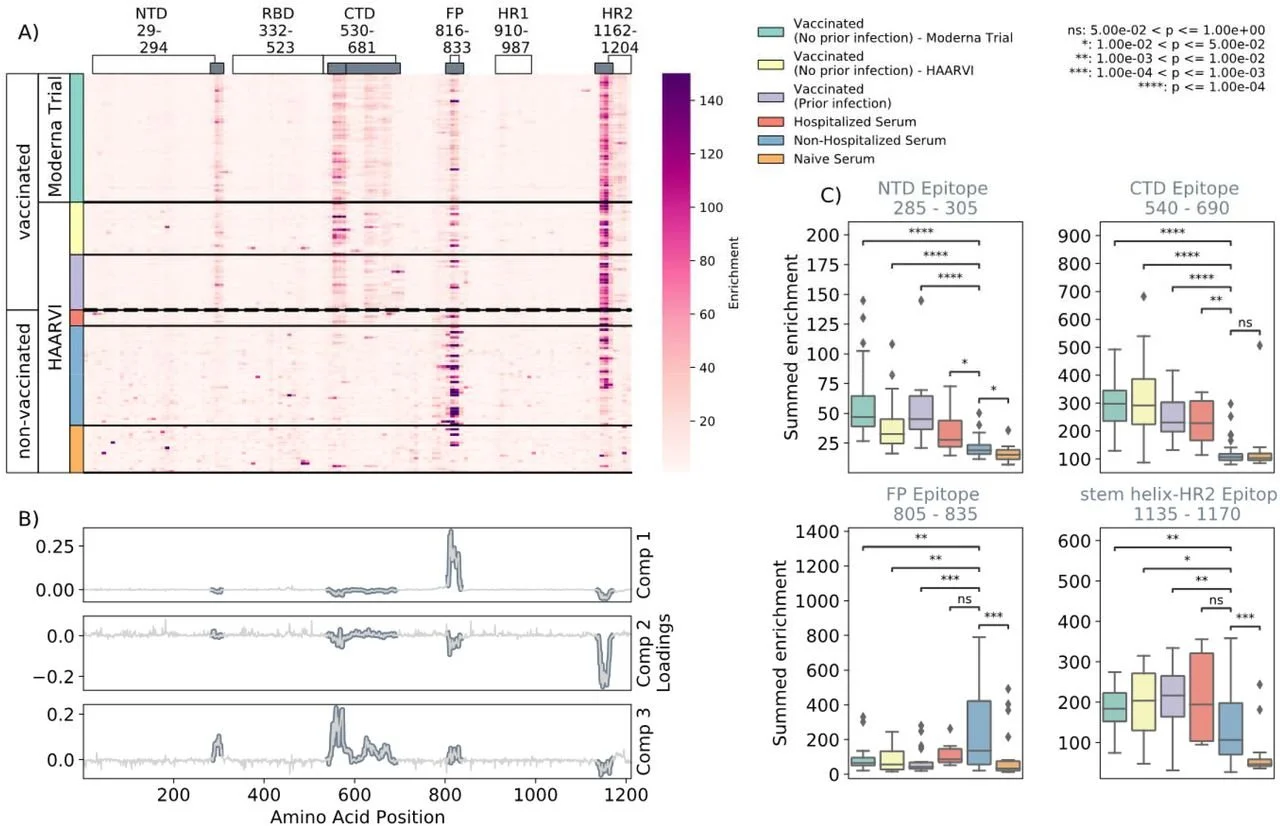
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein after infection and/or vaccination
In preprint on bioRxiv 10/7/2021
Vaccination is how many individuals will first acquire antibodies against SARS-CoV-2. Learning how vaccine acquired antibodies and infection acquired antibodies differ is important to potentially predict trends in antigen mutation. Researchers may be able to predict new variants by observing where antibodies bind to SARS-CoV-2 outside of currently known binding sites. Samples were obtained from participants in the Moderna Vaccine Trial and the HAARVI study. From these samples researchers were able to observe epitope binding sites, outside of the receptor-binding domain (RBD), from antibodies acquired from vaccination and antibodies acquired from infection. The RBD is a domain on the spike protein that SARS-CoV-2 uses to gain entry into host cells. Many of the vaccine-elicited antibodies specifically target the RBD. Having the ability to observe other epitope binding sites is helpful for researchers to learn more about how SARS-CoV-2 enters host cells, and for predicting where mutations may occur that could lead to vaccine elusive variants. The antibodies of those who were vaccinated and those who were hospitalized from SARS-CoV-2 infection had similar epitope binding sites (NTD, CTD, and SH-H regions) compared to those who had mild infection from SARS-CoV-2 (FP region). Factors such as vaccine dose, vaccine type and age were not found to affect how vaccine acquired antibodies bound to specific epitopes. Learning which epitopes antibodies bind to is important to help predict which parts of the pathogen’s genome may be vulnerable to mutation. This allows researchers to identify possible SARS-CoV-2 genome mutations that could lead to vaccine resistant strains.
DISCLAIMER: studies posted to medRxiv or biorxiv (med archive and bio archive) are pre-prints and have not been peer reviewed or published.
Dynamics of breast milk antibody titer in the six months following SARS-CoV-2 infection
Published in J Clin Vir 07/2021
This study analyzed breast milk samples from two lactating women post COVID-19 illness. It found a durable antibody response that began at 10 days post-symptom onset and persisted for up to 6 months after infection. These findings suggest that breast milk may play a role in immune protection of infants.
Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection
Published in Science Translational Medicine 6/30/2021
HAARVI samples were used in this study carried out by the Bloom lab at Fred Hutchinson Cancer Research Center. The study characterizes the differences between neutralizing antibodies produced by individuals who were naturally infected with SARS-CoV-2 and those produced by individuals who received Moderna’s mRNA vaccine. Antibodies created in response to the Moderna mRNA vaccine were more focused on the receptor binding domain (RBD) of the spike protein compared to those elicited by natural infection. These findings indicate that antibodies acquired through different modes of exposure, including different types of vaccines or natural infection, may produce different immune responses to SARS-CoV-2 and suggest some vaccine responses may be more susceptible to viral evolution.
High-resolution profiling of pathways of escape for SARS-CoV-2 spike-binding antibodies.
Published in Cell 05/27/2021
Antibody escape is an important area of research as the SARS-CoV-2 virus has continued to spread and new mutated variants emerge. Escape happens when specific areas of the spike protein on the surface of the SARS-CoV-2 virus mutate so antibodies are no longer able to bind to and inactivate the spike protein. If antibodies are not able to bind the mutant spike protein, the mutated virus can enter the host cell and our immune systems have to go through the process of making specific antibodies to the new spike protein from scratch.
The goal of this study was to characterize the possible mutations to the SARS-CoV-2 spike protein that would escape antibodies made to the wild-type (unmutated) virus. Mutated spike proteins were tested against samples of recovered individuals, provided by participants of the HAARVI study, to assess which mutations escape the most. They found that patterns of escape varied between individuals and that there were specific sites on the spike protein where mutations caused more escape. These findings can give insight into how potential variants could escape antibodies produced by natural infection or vaccines.
A differential regulatory T cell signature distinguishes the immune landscape of COVID-19 hospitalized patients from those hospitalized with other respiratory viral infections
Published in Science Advances 11/10/2021
Samples from the HAARVI study were used in collaboration with the Lund, Prlic, and Schiffer labs to compare the immune response between participants infected with Flu, RSV and SARS-CoV-2. They found a different immune response between the different viruses, especially looking at signalling proteins called interleukins (IL). The study also looked at the differences in response between healthy controls, moderate, severe and critically ill COVID-19 patients. There was a significant difference in the pro-inflammatory and T cell response of critically ill patients. This suggests that therapies targeting the regulatory T cell response could be useful in the treatment of severe COVID-19 disease.
Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies
Published in Cell Host & Microbe 03/10/2021
As emerging SARS-CoV-2 variants are discovered and are spreading around the world, very little is known regarding the risk these new mutants pose at infecting previously exposed and vaccinated individuals. In collaboration with scientists at Fred Hutch, this study deeply probed how changes in the Spike receptor-binding domain (RBD), the specific site that interacts with the ACE-2 receptor on human cells, could promote evasion from the protective neutralizing antibody pools of eleven previously infected individuals. The authors developed procedures that could detect which specific RBD mutations escaped antibody binding and neutralization within each individual. The authors identified three unique locations within RBD, where if mutations occurred, could dramatically reduce both antibody binding and neutralization. Mutations in one specific site, E484, significantly increased neutralizing antibody escape in some individuals, and such mutations have been identified in the South Africa and Brazil variants. However, the authors also observed extremely diverse antibody responses across the eleven participants, and some participants demonstrated potent neutralizing activity regardless of any mutation in RBD. The authors noted that this study investigated the RBD-specific neutralization activity in only eleven individuals, and understanding how antibody pools interact with the entire Spike antigen could greatly assist in future therapeutics. In summary, some mutations in the RBD, specifically at site E484, could reduce vaccine efficacy and drive reinfections, but site-specific antibody responses to SAR-CoV-2 are diverse, dynamic and require further elucidation.
Comorbid illnesses are associated with altered adaptive immune responses to SARS-CoV-2
Published in JCI Insight 02/23/2021
In collaboration with investigators at the Ragon institute and the University of Washington, the authors profiled the humoral and T-cell-specific immunity in hospitalized and non-hospitalized SARS-CoV-2 infected participants after recovery. The authors created thorough, individual immune fingerprints that included a wide range of antigen-specific antibody and cellular responses using convalescent sera and peripheral blood mononuclear cells (PBMCS) collected by the HAARVI study. The figure below summarizes the design of the study. Hospitalized individuals or those with comorbidities repeatedly demonstrated a robust but uncoordinated antibody and T-cell response to SARS-CoV-2 antigens. Basically, a strong but clumsy immune response during convalescence was greatly associated with participants who were very ill during acute infection. Conversely, non-hospitalized participants (moderate to mild symptoms) displayed a more coordinated but less dramatic immune profile during convalescence. Interestingly, the authors did not see a correlation between neutralizing antibody activity and illness severity which is contrary to many other studies, but the convalescent nature of these samples may have influenced these results. The authors speculated that the uncoordinated immune response and elevated risk of severe COVID-19 might be caused by a proinflammatory state more commonly observed in individuals with comorbidities such as diabetes and cardiovascular disease. Lastly, the data produced from this research could eventually be used to predict illness outcome early in infection and help guide the treatments against COVID-19.
Sequelae in Adults at 6 Months After COVID-19 Infection
Published in JAMA Network Open 2/19/2021
Many individuals who previously tested positive for COVID-19 have reported experiencing persistent symptoms for months after they’ve recovered from their illness, a phenomenon that has become known as long COVID or Post Acute Sequelae of COVID-19 (PASC). Our study team sought to assess the long term effects of COVID-19 among study participants. Between August and November 2020, HAARVI participants were asked to answer a follow-up questionnaire regarding persistent symptoms, additional medical care, changes in health related quality of life, and impacts on activities of daily living (ADLs). Of 177 participants who were at a median of 6 months post-infection, 30% reported at least 1 persistent symptom. The most commonly reported symptoms were fatigue and loss of sense of taste or smell. One-third of the cohort also reported a worsened quality of life compared to baseline, and 8% reported negative impacts on their Activities of Daily Living (ADLs). This work indicates that even relatively young, healthy individuals, including many who had only a mild case of COVID-19, may experience a host of symptoms that can last for months after infection and adversely impact quality of life. With millions of cases worldwide, even a small incidence of long term effects can have a huge impact on the economy and healthcare system.
This study has been covered by several media outlets including CNN, People, and HealthDay News. One of the lead authors on the study, Nick Franko, also recently wrote an op-ed regarding the impacts of long-covid that was published in the Seattle Times.
This figure shows the symptoms reported during the acute Covid-19 illness compared to those reported in the follow-up survey between 3 to 9 months post-infection. The results are stratified by illness severity.
Below is the questionnaire used in the JAMA Network Open publication. Please be aware that the questionnaire was administered in REDCap and branching logic does not appear in the images below.
Viral genome sequencing places White House COVID-19 outbreak into phylogenetic context
In preprint on medRxiv November 1, 2020
This study was published in collaboration with scientists at the Fred Hutchinson Cancer Research Center and there Brotman Baty Institute at the University of Washington. An event held at the White House Rose Garden in October 2020 was linked to the spread of over 50 COVID-19 cases. Two individuals associated with that event were enrolled in HAARVI and the genomes of the SARS-CoV-2 viruses from their samples were sequenced. The two samples were genetically very similar, and the two participants attested that they had no contact with each other leading up to their diagnoses and are therefore individually linked to the White House COVID-19 outbreak. When the sequences were compared to those of over 160,000 publicly available genomes, there were 5 distinct mutations that presented together only in the two samples collected for this study. The unique genetic identity of the virus that infected these individuals could help researchers trace downstream infections from that initial super-spreader event.
This figure shows the genetic diversity of SARS-CoV-2 samples collected from all over the world and indicates that the two samples from this study, designated as WH1 and WH2, fall within the viruses circulating as part of the US epidemic. Further analysis shows that they are descended from viruses circulating primarily in the US during March and April 2020.
Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2
Published in Cell October 30, 2020
As the COVID-19 pandemic continues, development of an effective vaccine is of the utmost importance. There are many different types of vaccines that use different strategies to give the recipient long-term and robust immunity. The researchers of this exhaustive study are exploring the development of a relatively new vaccine type where they create a small particle that displays only the exact protein needed for an effective immune response. In preclinical studies, similar vaccine candidates have been shown to create robust antibody responses against different targets such as HIV-1, RSV and influenza.
The team created a protein copy of the exact region of the SARS-CoV-2 virus that has been targeted by the immune system of individuals that effectively neutralized or destroyed the virus. For SARS-CoV-2, the region of focus is the receptor binding domain (RBD) of the spike protein that allows the virus to interact with and enter human cells. Next, they created a nano-particle that only displays approximately 60 copies of that protein, forcing the immune system to recognize only that structure and create antibodies against it. They then tested if mice injected with the nano-particles create antibodies that effectively neutralize the SARS-CoV-2 virus and compare the amount and structures of the antibodies produced to those of humans recovered from COVID-19. Serum from HAARVI participants was used as the comparison.
The results were very promising and suggest that a vaccine made with this technology could produce effective antibodies and therefore immunity in humans.
Epitope profiling reveals binding signatures of SARS-CoV-2 immune response and cross-reactivity with endemic HCoVs
In preprint on bioRxiv October 29, 2020
The authors sought to map which specific epitopes of SARS-CoV-2 antigens elicit antibody responses and whether certain epitopes are broadly recognized in mild/severe COVID-19 patients. The authors profiled epitope-specific antibody interactions by incubating sera from both SARS-CoV-2 infected and pre-pandemic healthy individuals with thousands of varying small, linear protein sequences that cover a large portion of the epitope repertoire from all seven human coronaviruses. By including all other human coronaviruses in this study, the authors were able to further probe for cross-reactive immune responses. Cross-reactive antibodies are evoked from previous coronavirus exposures and/or due to sequence similarity between the strains. The study discovered that despite considerable variability in the range of epitope-specific antibody responses in COVID-19 patients, many individuals share overlapping epitope activity. These broadly recognized epitopes can be targets for improved vaccine design and guide therapeutic drug development. Also, antibodies against regions of Spike, Nucleocapsid, and ORFab1 dominated the immunological responses in both mild and severe illness with very few responses to any other viral protein. These results coincide well with many similar studies and validate the results. Lastly, four cross-reactive epitopes were observed likely due to pre-pandemic coronavirus infections. Cross-reactivity in antibody responses can interfere with serological diagnostics and surveillance and drive negative immune outcomes. Greater understanding regarding which epitopes are cross-reactive will increase the efficacy of antibody testing and could provide insight into COVID-19-related immunity.
Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection
Published in The Journal of Infectious Diseases September 30, 2020
This study was published in collaboration with scientists at the Fred Hutchinson Cancer Research Center and examined the immune response over time of participants with COVID-19 and the difference in antibody response between individuals with varying levels of disease severity. The researchers found all study participants experienced a typical antibody response, with an initial peak in antibody levels shortly after infection followed by a decline as the individual recovered. Levels of neutralizing antibodies that targeted the SARS-CoV-2 virus declined modestly, with levels at three to four months post symptom onset generally about four-fold lower than those at one month. Most individuals still had substantial neutralizing antibody levels at three to four months after symptom onset and similar patterns were seen with several other types of antibodies that target the SARS-CoV-2 virus.
The study also found individuals who experienced more severe COVID-19 symptoms tended to have higher antibody levels shortly after infection but at 3 to 4 months post-symptom onset there was no significant difference in antibody levels between disease severity groups. Asymptomatic individuals experienced similar antibody responses to those who experienced mildly symptomatic COVID-19 illness episodes. The study findings show the immune response of most participants to COVID-19 was similar to a typical immune response seen with other known respiratory viruses. A dramatic decline in antibody levels after viral infections is very common as the body usually experiences a high production of short-lived antibody secreting cells while the individual is experiencing an active infection. For many other viral infections, antibody levels will decrease to a stable level that is then maintained for years or decades after infection. This stable level of antibodies has been associated with future immunity or improved illness outcome in case of reinfection with other viruses, and we may see similar results with COVID-19.
Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity
Published in Science September 29, 2020
This large, multi-institution collaboration examined samples from COVID-19 patients who experienced varying disease severity, in order to characterize the immune system’s response to the SARS-CoV-2 virus. They found that those infected with SARS-CoV-2 had a strong antibody response and that the antibodies produced by infected individuals targeted specific regions on the virus itself, primarily the spike protein and nucleoprotein. Blood samples collected before the COVID-19 pandemic had some antibodies that reacted mildly to the SARS-CoV-2 virus, but not to the extent of antibodies produced by COVID-19 patients nor to the specific SARS-CoV-2 proteins identified. This may be due to the frequent circulation of seasonal human coronavirus each year producing a mild immune response in most people. The study team also examined how the antibodies produced by individuals infected with SARS-CoV-2 reacted to other types of coronavirus. They found a strong reactivity to the SARS-CoV-1 virus (which caused an outbreak in 2003), and mild reactivity to more distantly related coronaviruses including several bat coronaviruses and seasonal human coronavirus.
The authors identified a difference in the antibody response between hospitalized and non-hospitalized participants, with hospitalized participants having a much stronger and broader antibody response to SARS-CoV-2, but also weaker antibody responses to prior infections from viruses that cause the flu and common cold. Hospitalized individuals were also more likely to have had two common types of herpes viruses, HSV-1 and CMV, which are known to have potential negative impacts on the immune system. The authors note it is difficult to determine if herpes infection impacted the severity of the SARS-CoV-2 infection or if differences in illness severity were influenced by demographic characteristics that are also associated with herpes prevalence, such as age, race and socioeconomic status. They also found that among hospitalized participants, males produced stronger SARS-CoV-2 antibody responses than females. Understanding the variations in immune response to SARS-CoV-2 infection is critical in the development of treatments and vaccines to combat the COVID-19 pandemic.
Experimental Drug Trial Utilizing HAARVI Samples
Article published in the NYT September 18, 2020
The New York Times recently reported on an ongoing experimental monoclonal antibody drug trial run by Eli Lily that one interviewed expert described as “rigorous, and the [preliminary] results are really compelling”. This study has not yet released any detailed data or been reviewed by independent experts, but we wanted to let our study community know samples from HAARVI contributed to the drug’s original development process. We’re hopeful this drug will prove to be effective once the data is released and published in a peer-reviewed journal and are excited about the contribution to any possible progress in developing treatments for COVID-19.
Distinct early serological signatures track with SARS-CoV-2 survival
Published in Immunity September 15, 2020
This paper was published in collaboration with scientists at the Ragon Institute of MGH, MIT, and Harvard. Researchers looked at specific antibody features of hospitalized SARS-CoV-2 patients, some of whom recovered and some of whom died. These antibody features were used to create a profile of each individual's immune response, and the immune responses of the individuals who survived were compared to those who died. This allowed researchers to explore whether the expression of certain antibody features could predict an individual’s disease outcome (death or recovery).
Researchers were able to identify five antibody-specific markers that collectively distinguished the two groups. Their findings indicated that individuals who recovered exhibited strong antibody responses to the spike protein found on the SARS-CoV-2 virus, while individuals who died had a stronger immune response to another protein found on the virus. These immune mechanisms could help steer the development of therapeutics and vaccine design and also help physicians discern which patients may need more medical attention or certain types of therapies.
Comparison of Unsupervised Home Self-collected Midnasal Swabs With Clinician-Collected Nasopharyngeal Swabs for Detection of SARS-CoV-2 Infection
Published in JAMA Network July 22, 2020
This study tested the effectiveness of people performing mid-nasal swabs on themselves at home to test for the presence of the SARS-CoV-2 virus. As states look to expand the amount of SARS-CoV-2 tests available, home-based testing strategies have been discussed as a way to help increase testing capacity and reduce risks to healthcare workers. An increase in at-home, self-collected swabs would help conserve limited personal protective equipment, such as gloves and masks, and allow individuals to get tested without leaving their home and potentially exposing themselves or other individuals to the virus.
To measure the effectiveness of the mid-nasal swabs, health care workers who were tested for SARS-CoV-2 infection using a clinician-collected nasopharyngeal swab also self-collected a mid-nasal swab at home, and the results from the two swabs were compared. The study findings suggest that self-collected nasal swabs are comparable to clinically collected swabs. In particular, the self-collected swabs had a sensitivity of 95% in individuals with high viral loads. High viral loads generally occur early in illness when risk of transmission is highest and people are less likely to seek medical care. These findings indicate that a home-based testing strategy offers a safe and scalable testing approach in a pandemic setting that can serve as a complement to clinician-collected nasopharyngeal swabs, especially when targeting individuals early in illness/immediately after symptom onset.
Analysis of a SARS-CoV-2 infected individual reveals development of potent neutralizing antibodies to distinct epitopes with limited somatic mutation
Published in Immunity July 14, 2020
This paper was published in collaboration with scientists at the Fred Hutchinson Cancer Research Center. The research team looked at how the immune system of an individual infected with SARS-CoV-2 responded to the virus, and where antibodies bound on the viral particles. These findings can give important insight into designing drugs and vaccines that could successfully destroy SARS-CoV-2 and even provide immunity against contracting the virus.
Scientists were specifically interested in finding neutralizing antibodies, which have the potential to prevent the virus from infecting new cells. The team found two neutralizing antibodies that were present in the blood 21 days after onset of clinical symptoms. The research team concluded that their results indicate that some powerful (high-affinity) coronavirus neutralizing antibodies require a short development period. Their findings suggest that a vaccine against the SARS-CoV-2 virus may only need to initiate a subset of B cells for potent neutralizing antibody responses to be manufactured. Pictured below is a figure that outlines the process the Fred Hutchinson research team utilized in their research study to identify potent neutralizing antibodies for potential therapeutic development.
Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein
Published in Nature Medicine July 10, 2020
A team at the Vanderbilt Vaccine Center is working on the isolation and characterization of monoclonal antibodies, which are known for their immunity against most RNA viruses, including SARS-CoV-2. Several groups in the U.S. are hoping to start clinical trials for COVID-19 monoclonal antibody treatments in the next 4-6 months.
The Vanderbilt team assigned monoclonal antibodies they discovered into five different groups based on their reactivity to the SARS-CoV-2 (S) spike protein. The team analyzed convalescent blood samples from four different subjects in North America that had recovered from COVID-19 during various time periods. The most potent monoclonal antibodies the Vanderbilt team discovered in their research could serve as biological candidates for preventing or treating SARS-CoV-2.
The figure above shows the process that the Vanderbilt team used for the rapid monoclonal antibody discovery outlined in their publication. Through discovery approach number 1 (on the left side of figure), they were able to find 310 monoclonal antibodies that reacted to the virus. Through the second approach (pictured on the right), 76 reactive monoclonal antibodies were found. Both workflows were performed in parallel, and the neutralizing assays used in the second discovery approach were completed in just 18 days, which is quite fast for the antigen sorting process. The team acknowledged that the timeline for isolating neutralizing antibodies can be limiting, as there is a need for maturation in memory B cells that are capable of neutralizing COVID-19.
Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual
In preprint on bioRxiv May 12, 2020
This paper was published in collaboration with researchers at the Fred Hutchinson Cancer Research Center. This study identified a variety of antibodies from a COVID-19 infected individual. A handful of these antibodies were identified to prevent and disrupt key processes used by the SARS-CoV-2 virus to infect human cells. The presence of antibodies could also indicate possible future immunity against reinfection of SARS-CoV-2.
The research team also analyzed the B cell response to the SARS-CoV-2 spike protein. The team was able to isolate monoclonal antibodies and characterize their neutralizing potency as well as the binding properties which indicate how well the antibodies were able to bind and neutralize the viral protein. The team concluded that the COVID-19 infected individual had developed a high amount of binding and neutralizing antibodies twenty-one days after infection.
Preliminary support for a 'dry swab, extraction free' protocol for SARS-CoV-2 testing via RT-qPCR
In preprint on bioRxiv April 23, 2020
This paper was published as a joint effort between research scientists at the University of Washington and the Brotman Baty Institute. The purpose of the study was to compare the effectiveness of a new, more direct, method of testing self-collected nasal swabs to the traditional method used to test for the SARS-CoV-2 virus. They developed and studied a simplified method of COVID-19 testing which reduces the need for certain supplies that are in high demand due to the current pandemic. A traditional nasal swab sample is transported in universal transport media (UTM) to ensure it is preserved and transported efficiently. The researchers were able to test the transport of a nasal swab without UTM, referred to as a dry swab, and eliminate the need to extract the RNA from a sample.
The figure above shows the range of detection of SARS-CoV-2 viral RNA with corresponding dry swabs (pictured in red and blue) and traditional UTM swabs (pictured in green) according to participant samples. The figure shows dry swabs perform very similar to traditional UTM swabs for RNA detection of SARS-CoV-2.
The results of the study suggest that the virus can still be detected with a dry swab sample collection. This could address the larger issue of the increasing demand for COVID-19 rapid testing as worldwide supply chains continue to struggle to meet this demand.
HAARVI Collaborators
To date our team has shared HAARVI samples with 16 different research groups at 12 institutions around the U.S. and Canada: